NCIMB 13751
NCIMB 13751
规格:
货期:
编号:TS391726
品牌:Testobio
| NCIMB number | NCIMB 13751 |
| Deposit type | Bacteria |
| Type strain | Yes |
| GMO | No |
| Taxon name | Aeromonas allosaccharophila |
| Depositor designation | Estere 289 |
| Preservation method | Lyophilised |
| Price band | A |
| Media | 233 |
| Gas regime | aerobic |
| Incubation period | 2 days |
| ACDP category | 2 |
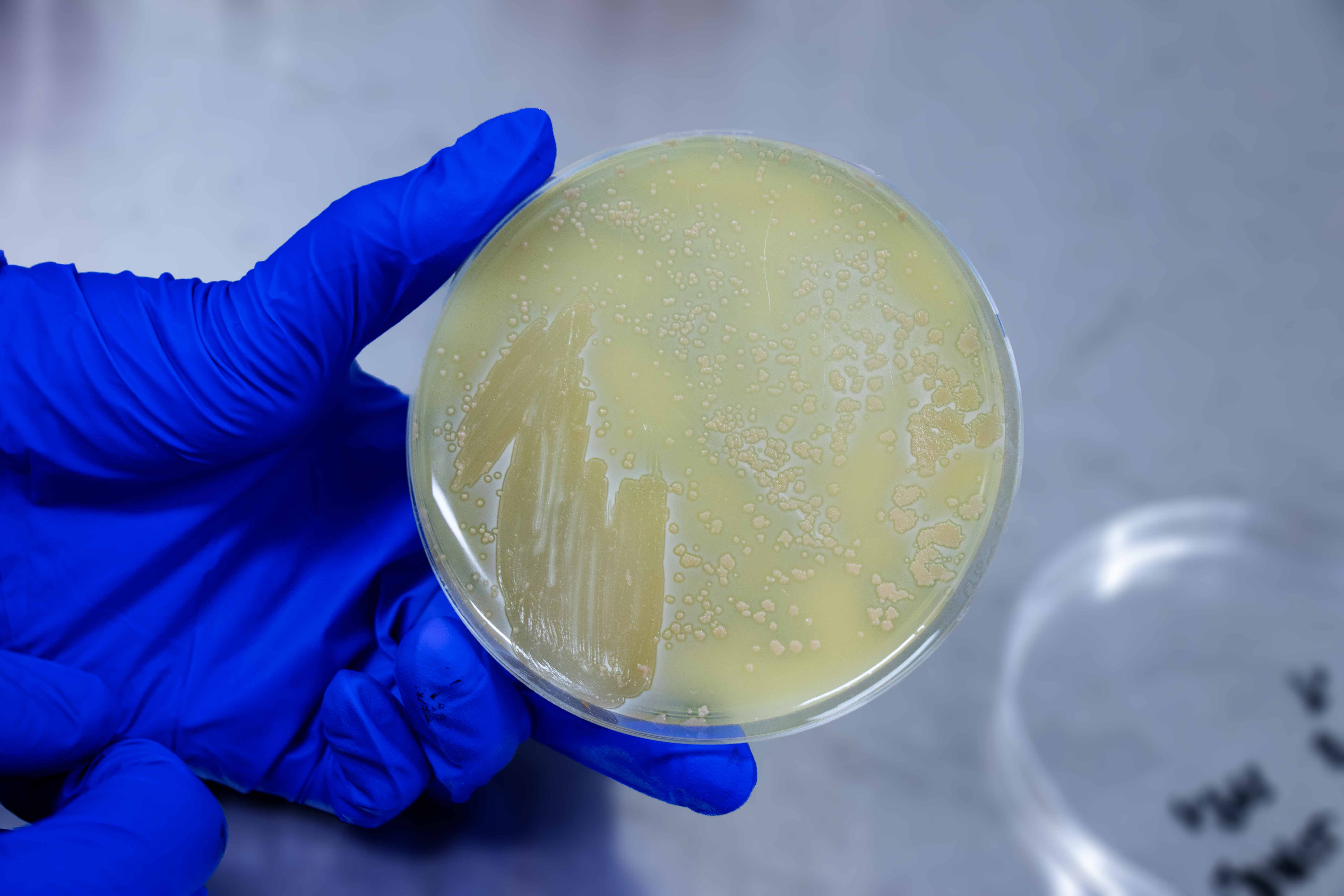
| Colony Edge | Entire |
| Colony Surface | Smooth |
| Colony Shape | Circular |
| Colony Elevation | Convex |
| Colony Colour | Off White |
| Cellular Shape | Rod |
| Gram stain | Gram Negative |
| Depositor Company | CECT |
| Depositor Address | |
| Source | "ex diseased elver of Auguilla auguilla (eel) Spain, Valencia, Polinya del Fucar fish farm" |
| Isolated by | C Esteve |
| Date of Accession | 30/10/2000 |
| Other collection IDs | ATCC51208 CCM4363 CCUG31218 CECT4199 DSM11576 |
| Yeast? | False |
| K12 | No |
| References | Martinez Murca et al IJSB (1992) 42: 511 FEMS Microb. Lett. (192) 91:199-206. (Unknown), FEMS Microbiol Lett 91, 199, 1992 Aeromonas allosaccharophila sp. nov., a new mesophilic member of the genus Aeromonas, FEMS Microbiol Lett 70(3), 199-205, 1992 Aeromonas aquariorum Is Widely Distributed in Clinical and Environmental Specimens and Can Be Misidentified as Aeromonas hydrophila, J Clin Microbiol 49(8), 3006-3008, 2011 Aeromonas in Finished Water by Membrane Filtration using Ampicillin-Dextrin Agar with Vancomycin (ADA-V).Washington, DC: Environmental Protection Agency, (journal unknown) , EPA EPA Method 1605, 2001 Determination of microbial diversity of Aeromonas strains on the basis of multilocus sequence typing, phenotype, and presence of putative virulence genes, Appl Environ Microbiol 77(14), 4986-5000, 2011 Evaluation of recA sequencing for the classification of Aeromonas strains at the genotype level, Lett Appl Microbiol 46(4), 439-444, 2008 Genetic relationships of Aeromonas strains inferred from 16S rRNA, gyrB and rpoB gene sequences, Int J Syst Evol Microbiol 56(12), 2743-2751, 2006 Malate dehydrogenase: A useful phylogenetic marker for the genus Aeromonas, Syst Appl Microbiol 33(8), 427-435, 2010 Multilocus genetics to reconstruct aeromonad evolution, BMC Microbiol 12(1), 62, 2012 Multilocus phylogenetic analysis of the genus Aeromonas, Syst Appl Microbiol 34(3), 189-199, 2011 Phylogenetic affiliation of Aeromonas culicicola MTCC 3249(T) based on gyrB gene sequence and PCR-amplicon sequence analysis of cytolytic enterotoxin gene, Syst Appl Microbiol 26(2), 197-202, 2003 Phylogenetic analysis and identification of Aeromonas species based on sequencing of the cpn60 universal target, Int J Syst Evol Microbiol 59(8), 1976-1983, 2009 Phylogenetic analysis of members of the genus Aeromonas based on gyrB gene sequences, Int J Syst Evol Microbiol 53(3), 875-883, 2003 Phylogenetic analysis of the genus Aeromonas based on two housekeeping genes, Int J Syst Evol Microbiol 54(5), 1511-1519, 2004 Ribosomal Multi-Operon Diversity: An Original Perspective on the Genus Aeromonas, PLoS One 7(9), e46268, 2012 Validation of the publication of new names and new combinations previously effectively published outside the IJSB. List No. 42, Int J Syst Bacteriol 42, 511, 1992 |
首页/产品中心/进口菌株
产品中心
联系我们 CONTACT US
0574-87917803
testobio@163.com
浙江省宁波市镇海区庄市街道兴庄路9号
创e慧谷42号楼B幢401室
最新促销
名称:NCIMB 13701
名称:NCIMB 13669
名称:NCIMB 13631
名称:NCIMB 13779
名称:NCIMB 13778
名称:NCIMB 13773
名称:NCIMB 13740
名称:NCIMB 13857
名称:NCIMB 13856
名称:NCIMB 13849
名称:NCIMB 13858
名称:NCIMB 13850

NCIMB 13751
NCIMB 13751
规格:
货期:
编号:TS391726
品牌:Testobio
标准菌株
定量菌液
DNA
RNA
规格:
冻干粉
斜面
甘油
平板
| NCIMB number | NCIMB 13751 |
| Deposit type | Bacteria |
| Type strain | Yes |
| GMO | No |
| Taxon name | Aeromonas allosaccharophila |
| Depositor designation | Estere 289 |
| Preservation method | Lyophilised |
| Price band | A |
| Media | 233 |
| Gas regime | aerobic |
| Incubation period | 2 days |
| ACDP category | 2 |
| Colony Edge | Entire |
| Colony Surface | Smooth |
| Colony Shape | Circular |
| Colony Elevation | Convex |
| Colony Colour | Off White |
| Cellular Shape | Rod |
| Gram stain | Gram Negative |
| Depositor Company | CECT |
| Depositor Address | |
| Source | "ex diseased elver of Auguilla auguilla (eel) Spain, Valencia, Polinya del Fucar fish farm" |
| Isolated by | C Esteve |
| Date of Accession | 30/10/2000 |
| Other collection IDs | ATCC51208 CCM4363 CCUG31218 CECT4199 DSM11576 |
| Yeast? | False |
| K12 | No |
| References | Martinez Murca et al IJSB (1992) 42: 511 FEMS Microb. Lett. (192) 91:199-206. (Unknown), FEMS Microbiol Lett 91, 199, 1992 Aeromonas allosaccharophila sp. nov., a new mesophilic member of the genus Aeromonas, FEMS Microbiol Lett 70(3), 199-205, 1992 Aeromonas aquariorum Is Widely Distributed in Clinical and Environmental Specimens and Can Be Misidentified as Aeromonas hydrophila, J Clin Microbiol 49(8), 3006-3008, 2011 Aeromonas in Finished Water by Membrane Filtration using Ampicillin-Dextrin Agar with Vancomycin (ADA-V).Washington, DC: Environmental Protection Agency, (journal unknown) , EPA EPA Method 1605, 2001 Determination of microbial diversity of Aeromonas strains on the basis of multilocus sequence typing, phenotype, and presence of putative virulence genes, Appl Environ Microbiol 77(14), 4986-5000, 2011 Evaluation of recA sequencing for the classification of Aeromonas strains at the genotype level, Lett Appl Microbiol 46(4), 439-444, 2008 Genetic relationships of Aeromonas strains inferred from 16S rRNA, gyrB and rpoB gene sequences, Int J Syst Evol Microbiol 56(12), 2743-2751, 2006 Malate dehydrogenase: A useful phylogenetic marker for the genus Aeromonas, Syst Appl Microbiol 33(8), 427-435, 2010 Multilocus genetics to reconstruct aeromonad evolution, BMC Microbiol 12(1), 62, 2012 Multilocus phylogenetic analysis of the genus Aeromonas, Syst Appl Microbiol 34(3), 189-199, 2011 Phylogenetic affiliation of Aeromonas culicicola MTCC 3249(T) based on gyrB gene sequence and PCR-amplicon sequence analysis of cytolytic enterotoxin gene, Syst Appl Microbiol 26(2), 197-202, 2003 Phylogenetic analysis and identification of Aeromonas species based on sequencing of the cpn60 universal target, Int J Syst Evol Microbiol 59(8), 1976-1983, 2009 Phylogenetic analysis of members of the genus Aeromonas based on gyrB gene sequences, Int J Syst Evol Microbiol 53(3), 875-883, 2003 Phylogenetic analysis of the genus Aeromonas based on two housekeeping genes, Int J Syst Evol Microbiol 54(5), 1511-1519, 2004 Ribosomal Multi-Operon Diversity: An Original Perspective on the Genus Aeromonas, PLoS One 7(9), e46268, 2012 Validation of the publication of new names and new combinations previously effectively published outside the IJSB. List No. 42, Int J Syst Bacteriol 42, 511, 1992 |